The dbBRCA-Chinese database is available at (https://dbbrca-chinese.fhs.um.edu.mo).
1. A new user registers with user’s name, email address and affiliated institution.
2. User logs in with email address and password to access the database.
3. User can choose to download the entire data set or to browse the database contents.
4. There are two browsing options: one is 'Browse and Search', the other is customized 'Browse and Filter'.
5. 'Search' tab allows users to input a keyword to search for a specific variant and its related information;
6. 'Filter' allows users to select gene, exon, method of variant detection, reported status, variant type and classification etc.
7. The variants are shown in the browsing table.
8. User can click the variant ID to open a new page for detailed information.
9. User can click the reference IDs to open a new page for reference information.
10. In the variants in the browsing table, the database also provides a direct link to view the selected variant in UCSC Genome browser.
11. 'Download' option allows to download the full data sets as excel files.
12. A mailing list ([email protected]) is set for database users and database developer to communicate database-related issues. Meanwhile, all the existing versions will remain accessible. Please refer to the publications for more details.
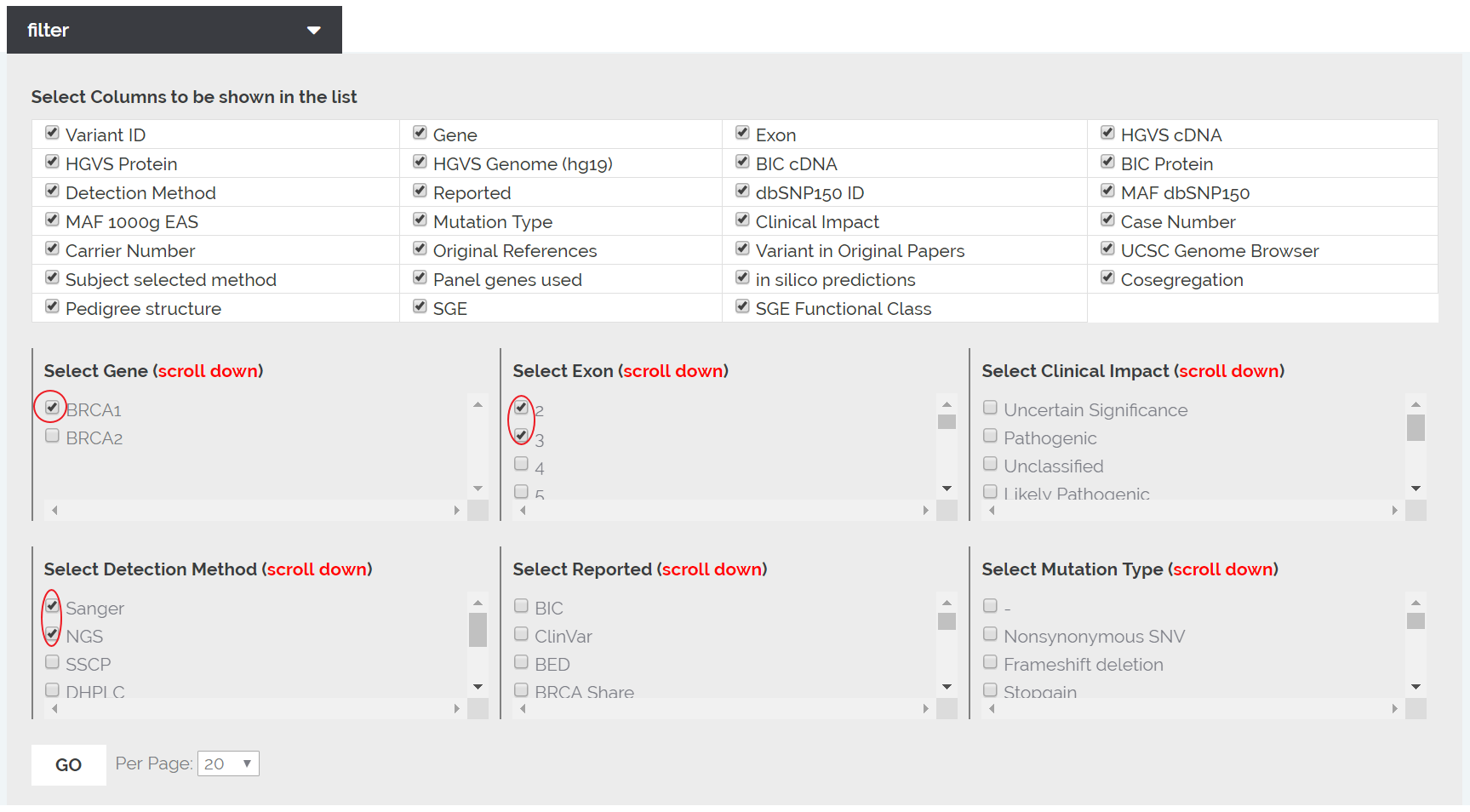
1. Register in the database.
2. Login to the database.
3. Click the 'Search and Filter' on the top navigation menu.
4. Click the box of 'BRCA1' In the 'Select Gene'.
5. Click the box of '2' and '3' in the 'Select Exon'.
6. Click the boxes of 'Sanger' and 'NGS' in the 'Select Detection Method'.
7. Optional, chose 20 at the 'Per Page' to show 20 lines per page.
8. Click 'GO' to search.
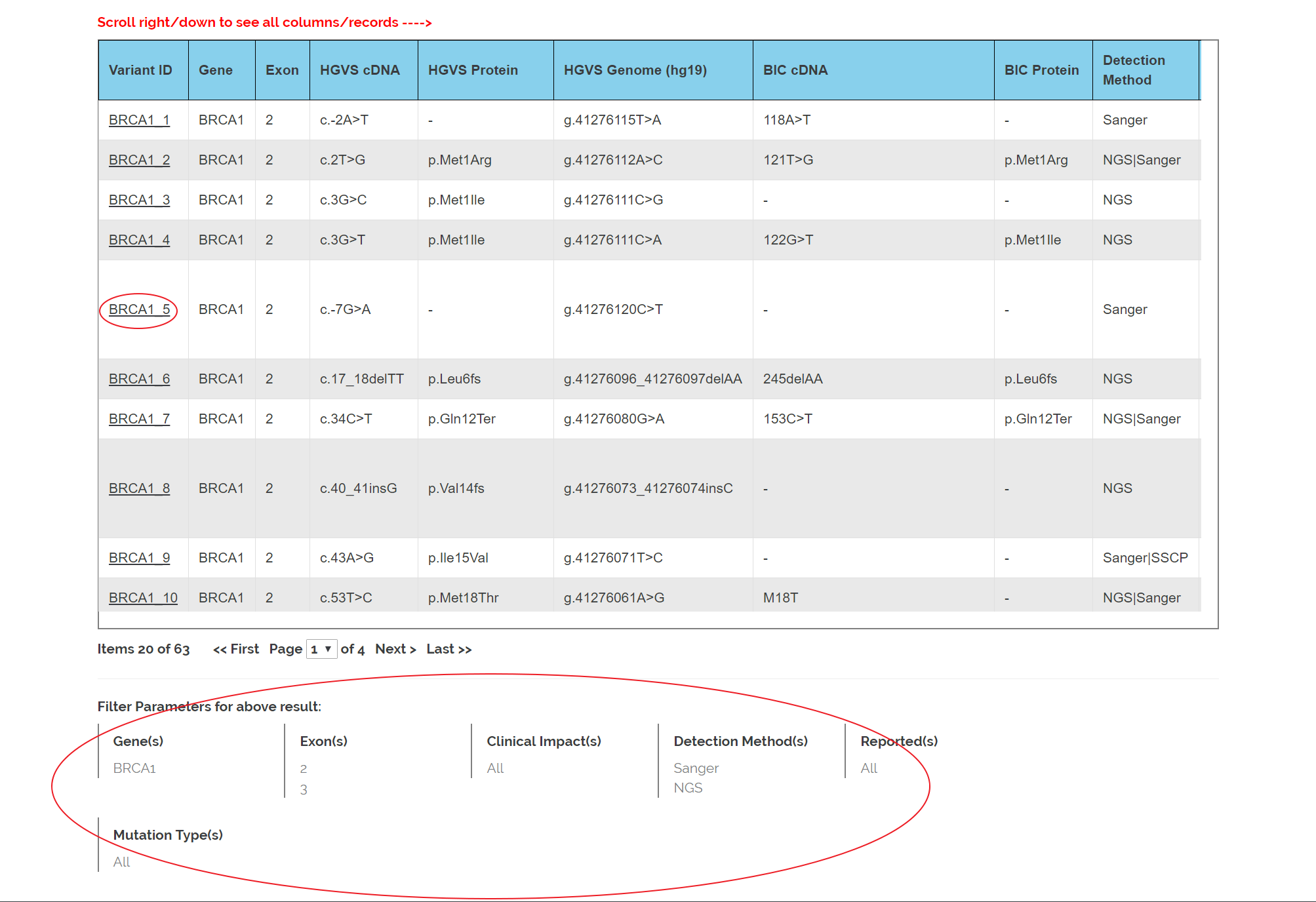
9. Variants in exon 2 and 3 of BRCA1 detected by Sanger and NGS are shown in the table.
10. Check the filter criteria shown at the bottom.
11. Click the variant ID to see detailed information. Detail information page please refer 'What is the detailed information of variants?' in 'Help'.
12. Click the Reference numbers to see references. Reference page please refer to 'What are the references?' in 'Help' and 'Use filtering to retrieve the data' in 'Help'.
13. Click the 'Genome Browser' to see variants in UCSC Genome Browser. Please refer to 'Use filtering to retrieve the data' in 'Help'.